Impact
The Plant Phenotyping Dataset and the CVPPP Challenges: 4 years later
This summer (June 2018) marks almost 4 years since we launched the first Leaf Segmentation Challenge and we made the first version of the dataset available. While we initially created a tech report to offer details on how we collected the data to accompany the challenge we quickly realized that making them open and publishing a paper that describes them such that it can remain in perpetuity it would be the best way to advance science forward.
Well we couldn't have been more right. In the following few paragraphs we want to briefly describe the current state of the data and the accompanying challenge in who uses them, who cites them, and how new efforts have been supported.
The data
The dataset appeared in the public domain in December 2015. Purposely we ask the users to fill in a form with some rudimentary information that we use to analyse how useful the data have been for the broad community prior to downloading the data. Our initial goal was to introduce the problem of phenotyping to mainstream computer vision. The analysis below draws from the data from this form.
Fig. 1. Close to exponential growth in downloads in the first three years (requests double every 12 months).
Fig. 2. Are we expanding beyond specialists that are familiar with plant phenotyping? Clearly yes. More than 60% of requests come from people that are not experts in the area (as of June 2018, please see figure for current numbers).
Fig. 3. Are users experts or also students? Almost 60% of the users identify themselves as students (as of June 2018, please see figure for current numbers).
Referring to Figures 1 to 3, some of the facts we can see:
- Exponential growth in requests to download (Fig. 1). At the time of writing this text (June 2018) more than 1300 requests were recorded.
- Approximately 70% of the requests originate from users that are not actually working in the problem already (Fig. 2).
- Almost 60% of users identify themselves as students of some capacity (Fig. 3). Remarkably 24% are undergraduates. Although it is not evident from the figures, 25% of the students are not working in agriculture related problems.
This even rudimentary analysis, demonstrates that we have accomplished our most important goal: that of introducing the societal important problem of plant phenotyping not only to the broad computer vision community but also directly to young researchers and students.
Impact in scholarly work
As of now (June 2018) the related paper has more than 35 citations according to Google Scholar. [1]
While some are peripheral mentions, or others come directly from the co-authors (can be seen as self-citations), a few really stand out as those are from colleagues publishing in top computer vision venues such as CVPR, and ICCV. This is not surprising since thanks to pioneering work from Romera-Paredes & Torr [2] and Ren & Zemmel [3] our dataset has become almost a benchmark in multi-instance segmentation. Dr Romera-Paredes (DeepMind), during his keynote at CVPPP @ICCV 2017 said, “The plant [CVPPP] dataset is considered the MNIST for multi-instance segmentation”. To be compared to MNIST, perhaps the most well-known dataset for image classification is definitely a strong indicator of the impact of this dataset.
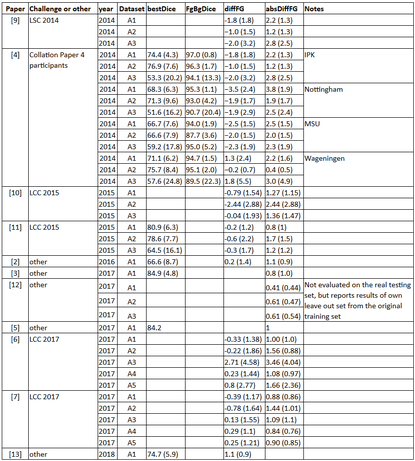
Table 1: Selected results of LSC and LCC so far (as of June 2018)
Several others have followed the trend of using the dataset to test and evaluate their pipelines. Table 1 shows the use of the dataset as of now. While we see that performance is improving, it seems that the broad computer vision community has focused on one source (A1) and as such we advise that more broad use and transfer learning attempts are made (i.e. to evaluate the performance of methods on other data sources as well A2 to A5).
Challenges on CodaLab
As a consequence of the success of the dataset and the frequent requests to evaluate results, we made the Leaf Segmentation Challenge (LSC) available on CodaLab recently. As of now 11 participants registered and submissions slowly reach the leaderboard. We hope that this service will be well received and further helps the community to transparently evaluate their findings.
Sources
[1] Google Scholar citationf for [8]
[2] Bernardino Romera-Paredes, Philip H. S. Torr, Recurrent Instance Segmentation, https://arxiv.org/abs/1511.08250
[3] Mengye Ren, Richard S. Zemel, End-To-End Instance Segmentation With Recurrent Attention. The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017, pp. 6656-6664
[4] H. Scharr et al.; Leaf segmentation in plant phenotyping: a collation study. Machine Vision and Applications, , Volume 27, Issue 4, pp 585–606,
[5] Bert De Brabandere, Davy Neven, Luc Van Gool, Semantic Instance Segmentation with a Discriminative Loss Function, https://arxiv.org/abs/1708.02551
[6] Shubhra Aich, Ian Stavness, Leaf Counting With Deep Convolutional and Deconvolutional Networks. The IEEE International Conference on Computer Vision (ICCV), 2017, pp. 2080-2089,
[7] Andrei Dobrescu, Mario Valerio Giuffrida, Sotirios A. Tsaftaris, Leveraging Multiple Datasets for Deep Leaf Counting. The IEEE International Conference on Computer Vision (ICCV), 2017, pp. 2072-2079
[8] M. Minervini, A. Fischbach, H.Scharr, and S.A. Tsaftaris. Finely-grained annotated datasets for image-based plant phenotyping. Pattern Recognition Letters, pages 1-10, 2015, doi:10.1016/j.patrec.2015.10.013
[9] Pape JM., Klukas C. (2015) 3-D Histogram-Based Segmentation and Leaf Detection for Rosette Plants. In: Agapito L., Bronstein M., Rother C. (eds) Computer Vision - ECCV 2014 Workshops. ECCV 2014. Lecture Notes in Computer Science, vol 8928. Springer, Cham
[10] Mario Valerio Giuffrida, Massimo Minervini and Sotirios Tsaftaris. Learning to Count Leaves in Rosette Plants. In S. A. Tsaftaris, H. Scharr, and T. Pridmore, editors, Proceedings of the Computer Vision Problems in Plant Phenotyping (CVPPP), pages 1.1-1.13. BMVA Press, September 2015.
[11] Jean-Michel Pape and Christian Klukas. Utilizing machine learning approaches to improve the prediction of leaf counts and individual leaf segmentation of rosette plant images. In S. A. Tsaftaris, H. Scharr, and T. Pridmore, editors, Proceedings of the Computer Vision Problems in Plant Phenotyping (CVPPP), pages 3.1-3.12. BMVA Press, September 2015
[12] Ubbens JR and Stavness I (2017) Deep Plant Phenomics: A Deep Learning Platform for Complex Plant Phenotyping Tasks. Front. Plant Sci. 8:1190. doi: 10.3389/fpls.2017.01190
[13] Amaia Salvador, Miriam Bellver, Victor Campos, Manel Baradad, Ferran Marques, Jordi Torres, Xavier Giro-i-Nieto, From Pixels to Object Sequences: Recurrent Semantic Instance Segmentation, https://arxiv.org/abs/1712.00617